|
|
Open, Save, Imports and Exports
The program uses a special format for data storage. JUICE files have the same name and three different suffixes (WCT - table data, colours, ecological informations about species etc.; EXP - header data; STR - header structure).
Displayed dialog box allows to select WCT files available in selected directory. Header data are only optional and needn't be imported and opened together with WCT file. They may be attached to WCT file additionally (see Import header data).
The table with all additional informations as colours, structure, species names, ecological values, separators etc. is saved through dialog box. The program will ask you to save current file before its end or selection of the function Open file.
Table and header data imports
The program is designed as an extension of the TURBOVEG database program. However, I have prepared some other import filters for users who have no possibility to use TURBOVEG. We have small experiences with these import gates and I will be pleasured if you will inform me about your success with them.
Import from TURBOVEG
Export table and header data from TURBOVEG:
Import table to the JUICE program
|
|
Connection of new relevés with an existing table
This function was created for an additional storage of new relevés into previously managed table. It is possible to connect only WCT table with some Cornell condensed file - the connection of two JUICE tables (WCT files) is not possible.
|
|
|
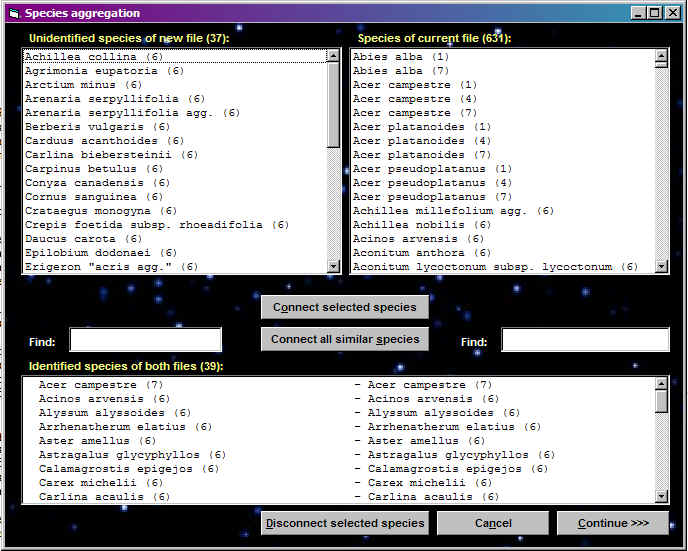 |
The structure of header data is selected in TURBOVEG and exported before the first import to JUICE. It may happen that some necessary information needed for the table analysis will miss after table preparation. This function allows to connect newly exported header data to an existing WCT file with new header informations.
|
|
Import from spreadsheed file (e. g. Microsoft EXCEL)
This table format is useful only for smaller tables. Please, for large tables use TURBOVEG database system or convert your table into Cornell condensed file. Study the table import format carefully!!!
|
|
This possibility allows to import data only with short cut species names. The structure of CC! file must be similar as in FILE IMPORT TABLE FROM TURBOVEG. Please, in problems contact author of the program.
It is the easiest import format for users without TURBOVEG. The table consists of four files with the same name and different suffixes: TXT, TAB, EXP, STR. There is an example of such table. These files can be also exported through EXPORT and TABLE as a text files.
TXT file consists of three columns (SPECIES NAME, LAYER, TABLE DATA) with the same number of characters which are separated by 5 spaces.
TAB file is optional. Each line contains a special identification number of relevé (a range from 1 to 999.999) as a TURBOVEG relevé number. The file can be omitted.
STR file defines table data (variable names) and their starting possition in EXP file.
EXP file contains relevé information
about the site and environmental factors. It must be created in accordance with
STR file.
|
 |
Short headers can contain only six characters per one relevé. They are displayed on the top ot the table and may contain Relevé number, Turboveg number, Sequence number, Group number, different counts or calculations (e. g. Equitability), part of header data (e. g. Year or Syntaxon code) or some other external information loaded through import of short headers.
|
The import allows to combine several informations into one header (For example it is possible to reserve first two characters for vegetation class and next four characters for a year of relevé sampling). The file have to contain Relevé number and short header value. The value can be copied only partly (define source range of copied characters) and can also replace only part of actual short head. Warrning!!! Please, before import of your data try to create exporting file with short headers and study its structure. |
|
Species data are stored near to species name in slightly grey field of the table. Species data can contain e. g. layer, some ecological or biological information about the species. This field can be used for species sorting and making separators among species groups. The import uses all text files with fixed lenght of lines, which contain a species name and species data information(s).
The JUICE program allows to export selected parts of tables, header data, latitude and longitude coordinates for the mapping program DMAP (Morton 2001), fidelity values for Interspecific associations of COCKTAIL group functions, Cornell condensed files, synoptic tables and average Ellenberg values. Generally, there are two export types. The first type uses ad hoc defined files, usually in *.TXT format. The second type uses *.RTF format into which you can make several exports of data of different type. This file remains open until you end your session or make a new selection under FILE and EXPORT and CURRENT FILE: <path and file name>.
The table is exported into an *.RTF file to facilitate editing and presentation.
Select relevé colour and mark the relevés to be exported
Check the *.RTF file setting (menu FILE EXPORT CURRENT FILE: <any path and file name>)
Press FILE EXPORT TABLE
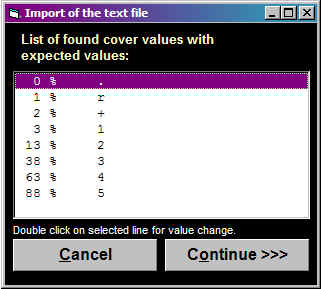
Choose colour of relevés to be exported and frequency threshold for species to be placed below the table.
If you want to change the
cover scale used in the output file, double-click in the
list box on any percentage value which needs a new
character value and change the output character.
The full synoptic table can be exported into the *.RTF file for editing and presentation.
Select relevé colour and mark the relevés to be exported
Check the *.RTF file setting (menu FILE and EXPORT and CURRENT FILE: <any path and file name>)
Display the table in the form of synoptic table (menu SYNOPTIC TABLE PERCENTAGE or CATEGORIES)
Press FILE EXPORT SYNOPTIC TABLE
Synoptic tables PERCENTAGE, CATEGORIAL and FIDELITY VALUE can be saved into the selected RTF file and replaced into some text editor for final editing. Before export display the synoptic table in required form and then select FILE EXPORT SYNOPTIC TABLE. The synoptic table will be added into the current file (menu FILE EXPORT CURRENT FILE: <file name>).
Table export in Cornell condensed format
A table modified by JUICE can be exported into a Cornell condensed file which is an input file for several other applications.
Select relevé colour and mark relevés to be exported
Press FILE EXPORT CC! FILE FOR (colour) RELEVÉS and save the
file.
The MATLAB export was developed for neural network data analysis. Please, try to study the structure of exported file or contact author of the program.
MULVA software is DOS classification package which is freely available on internet. The program JUICE supports export and it is possible to sort table according to program results.
This function creates a text file with selected items. For example, latitude/longitude export can be used for authomatic distribution mapping of relevés belonging to the same group.
Select relevé colour and mark relevés to be exported
Press FILE EXPORT HEADER DATA FOR (colour) RELEVÉS
Select header items to be
exported (use mouse and CTRL or Shift button) and save
the file.
The JUICE program can create the *.DIS and *.NAM files necessary for mapping with DMAP- (Morton 2001). There are three ways how to do this: (1) export species (and also saved species groups) of a selected colour, (2) relevés of a selected colour or (3) export the entire table. All variants of DMAP export are available in the menu FILE EXPORT DMAP FILE.
Warrning!
The program is possible to run DMAP automatically in the case you
have DMAP installed in the directory C:\DMAPW.
Short headers can contain only six characters per
one relevé. They are displayed on the top ot the table and may contain Relevé
number, Turboveg number, Sequence number, Group number, different counts or
calculations (e. g. Equitability), part of header data (e. g. Year or Syntaxon
code) or some other external information loaded through import of short headers.
The file is exported into a text file, which can be used e. g. as an import to
Access (for cummulation of results).
Species data are stored near to species name in
slightly grey field of the table. Species data can contain e. g. layer, some
ecological or biological information about the species. This field can be used
for species sorting and making separators among species groups. The export
cleate a text file with defined fixed lenght of each line. The file contains
species name and species data. Each exported file with species data can be
imported through function Species data import.
Export of all interspecific associations
Interspecific association (expressed as species-to-species fidelity value) can be calculated for all pairs of species present in the data set. Selected species and a list of the most associated species, sorted by decreasing fidelity, can be exported into *.RTF file. This procedure is repeated for all species in the data set whose frequency is higher than selected threshold. An example is stored at the web page: http://www.sci.muni.cz/botany/assoc_a.htm.
Select relevé colour and mark relevés to be exported
Check the *.RTF file setting (menu FILE EXPORT CURRENT FILE: <path and file name >)
Press FILE EXPORT ALL INTERSPECIFIC ASSOCIATIONS
Select the number of associated species to be exported and the frequency threshold for species to be considered. If you want to export all values set the first column to the maximum and second to the minimum value possible. The program will calculate interspecific associations for all species and export the species meeting the selected criteria, along with their fidelity values.