COCKTAIL
classification
- COCKTAIL
algorithm for defining species groups
The COCKTAIL algorithm (Bruelheide
1995, 2000) was designed for statistical forming of sociological
species groups. It proceeds iteratively as follows:
Step 1. The
algorithm can be used either to 1a – select species
differential for a vegetation unit, or 1b – define a vegetation
unit characterised by a group of differential species.
Step 1a. Starting
with preselected relevés (typical of a known vegetation unit)
the algorithm begins by calculating all species fidelities to
that vegetation unit and takes the species with the highest
fidelity values as the starting species group.
Step 1b. Starting
with a preselected species group which is user-defined, based on
literature or previous analysis.
Step 2. The
number of species of the species group is calculated in each
relevé. The expected and observed cumulative distribution
functions for relevés having 0 to k species are calculated. The
distributions’ intersection defines the required minimum number
m of species for a relevé to belong to the vegetation unit. The
vegetation unit is defined by all relevés having m or more
species belonging to the species group. If there is no
intersection between observed and expected cumulative
distribution then the algorithm aborts. This is the case when
species having fewer co-occurrences than expected form the
starting group.
Step 3. The
occurrences of each species in the vegetation unit are counted
and the fidelity is calculated.
Step 4. For all
species in the species group fidelity value is tested against an
(initially) fixed threshold fidelity. If fidelity exceeds the
threshold the algorithm proceeds to step 5. If not there are two
possibilities:
Step 4a. One of
the initially selected species does not exceed the threshold. The
group is rejected and the algorithm aborts.
Step 4b. The last
species added has caused a previous species’ fidelity to
decrease below the threshold. The previous species is removed,
and the algorithm does not try to add this species again until
the group has been changed by adding a further species.
Step 5. All
species not belonging to the species group are sorted according
to their fidelity value. If any exceed the threshold fidelity the
algorithm proceeds to step 6. If not the algorithm stops. The
species group is optimised when all species above the threshold
are included.
Step 6. The
species group is enlarged by including the (single) species with
highest fidelity. Iteration continues at step 2.
Note that step 4a guarantees that
the species group composition is not changed to such a degree
that the initial species no longer have the highest fidelity.
This restriction allows the formation of a number of species
groups, some with lower maximal fidelity than others. Not every
such group can be optimised. This is the case if species which do
not co-occur with the vegetation type more than expected form the
starting group.
When starting with preselected
relevés (belonging to a known syntaxon) the vegetation unit is
optimized in such a way that it is defined by differential
species groups a posteriori, and the final composition of relevés
in the group may be different than at the beginning. Not all
syntaxa can be defined by groups of differential species – some
are defined by dominance rather than by floristic composition.
Contrary to the original
description of the COCKTAIL algorithm, JUICE allows the user to
be more directly involved into the process of species group
formation. Instead of automatical checking the species group
composition in each step against the initially fixed fidelity
value, it can be checked by the user if its give sense in
phytosociological terms.
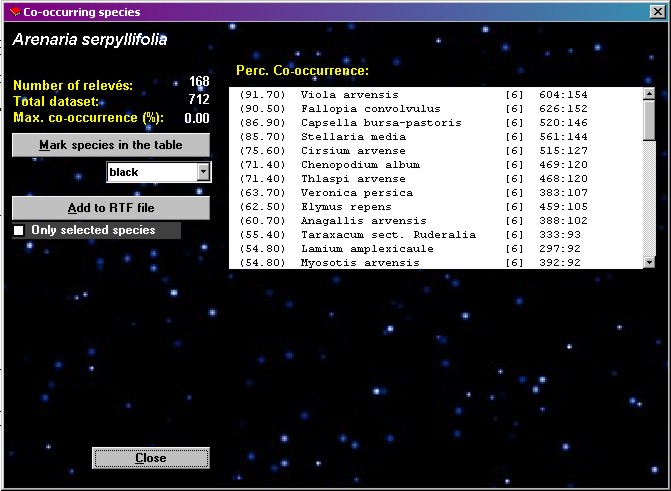
Co-occurring
species
| This function
finds species, which are most frequent in the relevés
where selected species occurs. The species must be
selected by mouse click in the Species table part before
the running the function. The function is called from the
menu ANALYSIS and CO-OCCURRING SPECIES. The selected species is shown at
the top of the Co-occurring species window, with the
number of relevés in which it occurs below. The list of
the most frequently co-occurring species is sorted by
decreasing frequency in the relevés where the selected
species occurs. The value shown in the first column is
the percentage of relevés of the selected species also
containing the listed species. The next columns are:
species name, layer, species frequency in the dataset,
and frequency of joint occurrence of current and selected
species.
|
|
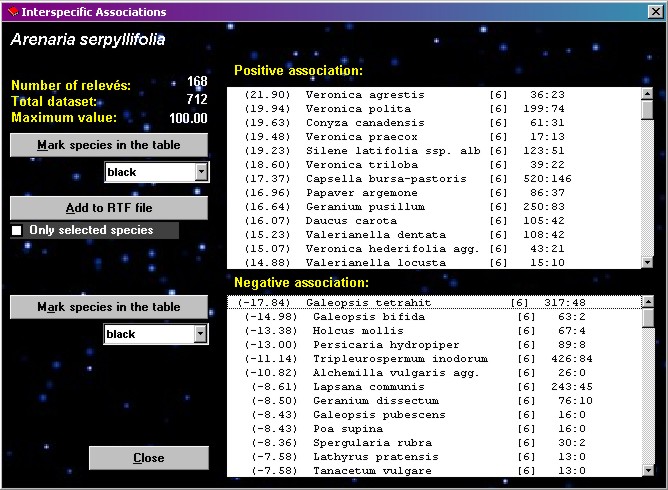
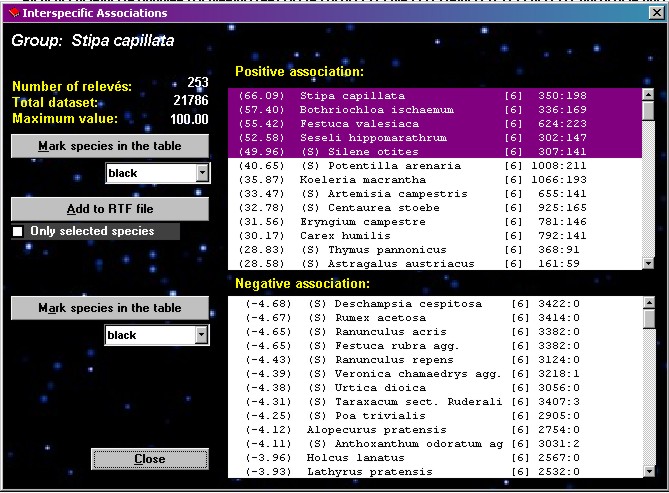
Interspecific
associations
| This
procedure is the basic step of COCKTAIL method; it tests
interspecific association between the selected species
and each other species in the table. A fidelity measure
is calculated for each pair of species, which gives
information on their reciprocal affinity in the dataset.
Fig. 17 shows the display form of this function, with a
sorted list of species positively and negatively
associated with the species previously selected by
clicking in the Species part of the table. The list with
positive associations can be exported to the current
export *.RTF file (see Section 10). All species in the list box can be
marked by shift or control button and mouse click by
using the button MARK SELECTED SPECIES IN THE TABLE.
The function INTERSPECIFIC
ASSOCIATIONS is also used in the other parts of the
program – Export of all interspecific associations (Section
10.3), Calculation of indicator values for species II (Section
7.4), Dependence sorting (Section 6) and INI groups (Section
9.5).
Explanation of list
columns: fidelity measure, species name, layer, species
frequency in the data set, frequency of joint occurrence
of current and selected species in the data set.
|
 |
An example of
difference between mentioned functions
The maximum value (100 %) in the
function Co-occuring species has a comparison of the species A
with both B and C, while the function Interspecific associations
gives a maximum fidelity value only for comparison of species A
with C.
- Species A
++++++++++..........................
- Species B
++++++++++++++++++++++++++..........
- Species C
++++++++++..........................
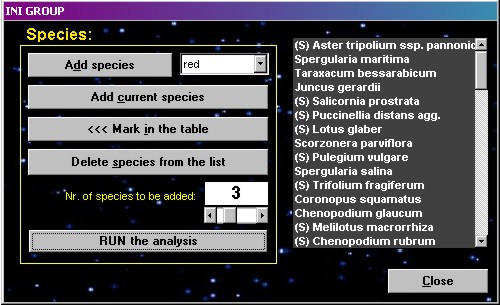
INI groups
| This function
searches for an appropriate species combination as a
starting group for the function COCKTAIL groups, i. e. a
group of two or more species which frequently occur
together. Select one or a few species and add them into
the form. State the number of species to be added to the
group (1-10 can be added at a time) and press the Run
button. The function calculates interspecific
associations of the first species from the list with all
the other species, sorts them by decreasing values, and
selects the one or more most associated species. These
most associated species are added to the list. The
procedure is repeated with the second species and with
all subsequent species now in the list, and is terminated
after testing all listed species. Mark backwards in the
table marks the listed species with the specified colour. Warning!
High number of species to be added in larger tables can
cause a very long cycle, so the program will have to be
manually interrupted.
|
 |
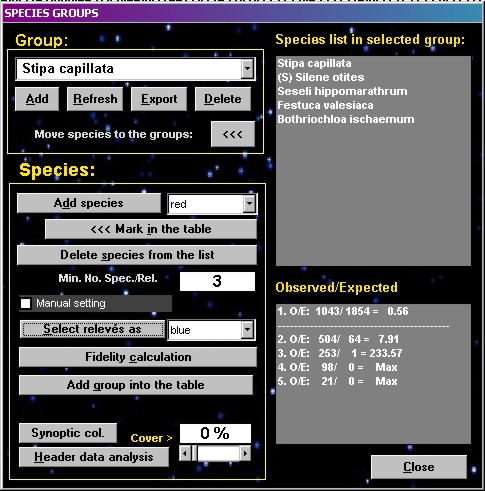
COCKTAIL
groups
| The function
COCKTAIL GROUPS searches for optimal combinations of
species, which have similar distributions in the data set
and can be used as sociological species groups in
vegetation classification. The reciprocal test of species
associations is based on the fidelity calculation. A
relevé is considered to contain the species group if
more than half of species of the group occur in it. How to create a species
group:
- Select a starting
group of species (one or more species, which were
selected by the functions Interspecific
associations or INI groups); all the selected
species should have the same colour.
- Open the form and add
starting group into the list box.
- Calculate the minimum
number of selected species which must be present
in a relevé if the group is to be considered
present in it. The minimum is calculated
statistically so that observed frequency of joint
occurrence of several species exceeds the
expected frequency in case of random and
independent distribution of these species in the
dataset. However, using half of the selected
species as the minimum value is better in many
cases. The desired minimum value can be specified
manually after the calculation of statistical
minimum value is done.
- Recolour relevés
where at least the minimum number of species is
present (i.e. where the group is present).
- Calculate fidelity
for the coloured group of relevés – a new form
displays.
- Select any new
species, which has similar distribution.
- Recolour this species
and add it into the COCKTAIL groups form.
- Continue from point 3.
Terminate the process when
the group seems to be optimal for syntaxonomic
classification. This may be when it is similar to a group
of diagnostic species traditionally recognized in the
syntaxonomic literature.
|
|
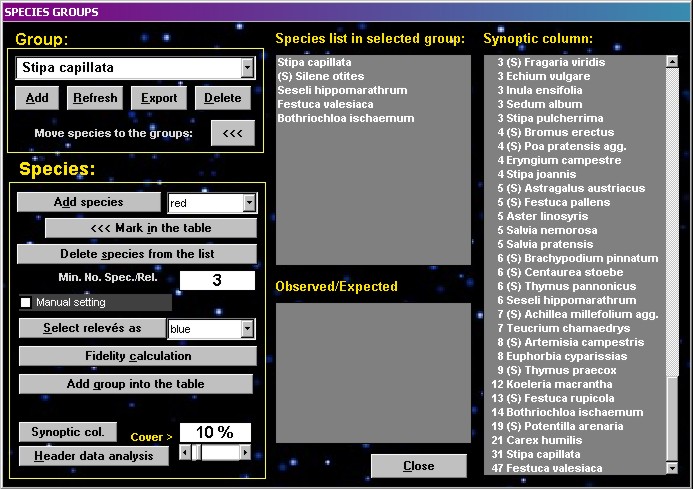
An optimized group with relatively
high fidelity values (see bellow) can be saved by writing its
name into the combo box and using the function ADD GROUP INTO THE
TABLE. If you wish to see the percentage synoptic column only for
coloured relevés, select the SYNOPTIC COL button.
Species groups can be added
directly into the table as a pseudospecies (with ### before the
group name). Such groups can be treated and analysed in the same
way as proper species and can be combined with other species in
the function Group aggregation. The column defined by selected
species group can be tested for constants and dominants displayed
after setting of cover threshold parameter (Header data analysis).
Group
aggregation
The group aggregation function
selects relevé groups by combining presence of species groups
and dominance of individual species. It uses species groups
loaded in the table and dominant species which are defined by
cover values exceeding a selected threshold. Species groups and
dominant species are combined by the logical operators AND, OR,
AND NOT, with the hierarchy defined by parentheses.
The query contains logical
operators AND, OR and NOT (= AND NOT). Species group names
consist of the characters ### followed by the species group name.
Names of dominant species are not preceded by characters ###, but
they have suffixes such as UP05 or UP25. For example, UP05 means
that species is considered if its cover in the given relevé is
higher than 5 % (UP25 means higher than 25 %).
Before running the query it can be
checked by Show definition.
Warning:
All pairs of logical variables associated by one operator must be
put in parentheses!
For details of the procedures
described in Sections 9.6 and 9.7, see Bruelheide (1995, 1997,
2000) or Bruelheide & Jandt (1995).
Expert
system
Expert system can automatically
assign a relevé to a vegetation type, if there is already a
classification based on species groups. The classification
algorithm must be included in *.ESY file. This file should
preferably be created as a product of the classification in a
large dataset and must include all required information on
aggregated species, species groups and their combinations into
vegetation types. A result of the expert system run is shown in
Fig. 21. The *.ESY file has a text structure and can be created
manually.
- In the first part
of this file aggregated species are defined:
- (example – species name;
species number; number of species aggregated; Turboveg
numbers of aggregated species)
-
- (S) Allium senescens ssp. m;47972;2;456;457;
- (S) Artemisia campestris;47981;2;860;861;
- (S) Avenula pratensis;47985;2;1183;1184;
- -1
- (the end of this section is
marked by -1)
-
- The next section
defines species groups:
- (example – group name;
minimum number of species in one relevé; Turboveg
numbers of species included.......)
-
- Agrimonia eupatoria;4;210;1096;2954;11496;
- Asplenium cuneifolium;3;1038;11236;838;
- Astragalus austriacus;4;1088;2645;12080;1101;
- -1
- (the end of this section is
also marked by -1)
-
- The last section
contains logical formulas with community definitions:
- (First line – community
name, second line – logical formula)
-
- Asplenium cuneifolium-Sesleria
albicans community
- <### Asplenium
cuneifolium>AND(<### Sesleria
albicans>OR<Sesleria albicansUP25>)
- Cirsium pannonicum-Sesleria
albicans community
- [<Sesleria albicansUP05>AND<###
Cirsium acaule>]NOT[(<### Sesleria
albicans>OR<Sesleria albicansUP50>)OR<###
Festuca pallens>]
-





